- Record: found
- Abstract: found
- Article: found
The Hemiparasitic Plant Phtheirospermum (Orobanchaceae) Is Polyphyletic and Contains Cryptic Species in the Hengduan Mountains of Southwest China
Read this article at
Abstract
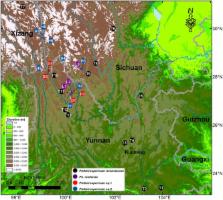
Phtheirospermum (Orobanchaceae), a hemiparasitic genus of Eastern Asia, is characterized by having long and viscous glandular hairs on stems and leaves. Despite this unifying character, previous phylogenetic analyses indicate that Phtheirospermum is polyphyletic, with Phtheirospermum japonicum allied with tribe Pedicularideae and members of the Ph. tenuisectum complex allied with members of tribe Rhinantheae. However, no analyses to date have included broad phylogenetic sampling necessary to test the monophyly of Phtheirospermum species, and to place these species into the existing subfamiliar taxonomic organization of Orobanchaceae. Two other genera of uncertain phylogenetic placement are Brandisia and Pterygiella, also both of Eastern Asia. In this study, broadly sampled phylogenetic analyses of nrITS and plastid DNA revealed hard incongruence between these datasets in the placement of Brandisia. However, both nrITS and the plastid datasets supported the placement of Ph. japonicum within tribe Pedicularideae, and a separate clade consisting of the Ph. tenuisectum complex and a monophyletic Pterygiella. Analyses were largely in agreement that Pterygiella, the Ptheirospermum complex, and Xizangia form a clade not nested within any of the monophyletic tribes of Orobanchaceae recognized to date. Ph. japonicum, a model species for parasitic plant research, is widely distributed in Eastern Asia. Despite this broad distribution, both nrITS and plastid DNA regions from a wide sampling of this species showed high genetic identity, suggesting that the wide species range is likely due to a recent population expansion. The Ph. tenuisectum complex is mainly distributed in the Hengduan Mountains region. Two cryptic species were identified by both phylogenetic analyses and morphological characters. Relationships among species of the Ph. tenuisectum complex and Pterygiella remain uncertain. Estimated divergence ages of the Ph. tenuisectum complex corresponding to the last two uplifts of the Qinghai–Tibet Plateau at around 8.0–7.0 Mya and 3.6–1.5 Mya indicated that the development of a hot-dry valley climate during these uplifts may have driven species diversification in the Ph. tenuisectum complex.
Related collections
Most cited references79
- Record: found
- Abstract: not found
- Article: not found
Inference from Iterative Simulation Using Multiple Sequences
- Record: found
- Abstract: found
- Article: not found
SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information
- Record: found
- Abstract: found
- Article: not found