- Record: found
- Abstract: found
- Article: found
An LC-MS Approach to Quantitative Measurement of Ammonia Isotopologues
Read this article at
Abstract
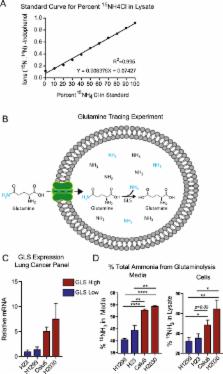
Ammonia is a fundamental aspect of metabolism spanning all of phylogeny. Metabolomics, including metabolic tracing studies, are an integral part of elucidating the role of ammonia in these systems. However, current methods for measurement of ammonia are spectrophotometric, and cannot distinguish isotopologues of ammonia, significantly limiting metabolic tracing studies. Here, we describe a novel LC-MS-based method that quantitatively assesses both 14N-and 15N-isotopologues of ammonia in polar metabolite extracts. This assay (1) quantitatively measures the concentration of ammonia in polar metabolite isolates used for metabolomic studies, and (2) accurately determines the percent isotope abundance of 15N-ammonia in a cell lysate for 15N-isotope tracing studies. We apply this assay to quantitatively measure glutamine-derived ammonia in lung cancer cell lines with differential expression of glutaminase.
Related collections
Most cited references14
- Record: found
- Abstract: found
- Article: not found
Ammonia derived from glutaminolysis is a diffusible regulator of autophagy.
- Record: found
- Abstract: found
- Article: not found
Engineering the gut microbiota to treat hyperammonemia.
- Record: found
- Abstract: found
- Article: not found