- Record: found
- Abstract: found
- Article: found
Characterization of oral and cloacal microbial communities of wild and rehabilitated loggerhead sea turtles ( Caretta caretta)

Read this article at
Abstract
Background
Microbial communities of wild animals are being increasingly investigated to provide information about the hosts’ biology and promote conservation. Loggerhead sea turtles ( Caretta caretta) are a keystone species in marine ecosystems and are considered vulnerable in the IUCN Red List, which led to growing efforts in sea turtle conservation by rescue centers around the world. Understanding the microbial communities of sea turtles in the wild and how affected they are by captivity, is one of the stepping stones in improving the conservation efforts. Describing oral and cloacal microbiota of wild animals could shed light on the previously unknown aspects of sea turtle holobiont biology, ecology, and contribute to best practices for husbandry conditions.
Results
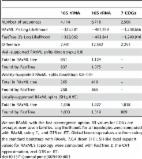
We describe the oral and cloacal microbiota of Mediterranean loggerhead sea turtles by 16S rRNA gene sequencing to compare the microbial communities of wild versus turtles in, or after, rehabilitation at the Adriatic Sea rescue centers and clinics. Our results show that the oral microbiota is more sensitive to environmental shifts than the cloacal microbiota, and that it does retain a portion of microbial taxa regardless of the shift from the wild and into rehabilitation. Additionally, Proteobacteria and Bacteroidetes dominated oral and cloacal microbiota, while Kiritimatiellaeota were abundant in cloacal samples. Unclassified reads were abundant in the aforementioned groups, which indicates high incidence of yet undiscovered bacteria of the marine reptile microbial communities.
Conclusions
We provide the first insights into the oral microbial communities of wild and rehabilitated loggerhead sea turtles, and establish a framework for quick and non-invasive sampling of oral and cloacal microbial communities, useful for the expansion of the sample collection in wild loggerhead sea turtles. Finally, our investigation of effects of captivity on the gut-associated microbial community provides a baseline for studying the impact of husbandry conditions on turtles’ health and survival upon their return to the wild.
Related collections
Most cited references69
- Record: found
- Abstract: found
- Article: not found
DADA2: High resolution sample inference from Illumina amplicon data
- Record: found
- Abstract: not found
- Article: not found
Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2
- Record: found
- Abstract: found
- Article: found