- Record: found
- Abstract: found
- Article: found
The DsrD functional marker protein is an allosteric activator of the DsrAB dissimilatory sulfite reductase
Read this article at
Significance
Metagenomic data have recently transformed our view of the role played by sulfur metabolism in anoxic environments by showing that this trait is much more widespread than previously believed. A key enzyme in sulfur metabolism is the dissimilatory sulfite reductase DsrAB that is ubiquitous in organisms with a reductive, oxidative, or disproportionating activity. However, the function of some dsr genes, such as dsrD, has so far been unknown despite its use as a functional marker to genomically assign the type of sulfur energy metabolism, sometimes with unclear results. Here, we disclose the function of DsrD as an activator of DsrAB that significantly increases its activity, providing important insights into the mechanism of this enzyme in different types of sulfur metabolism.
Abstract
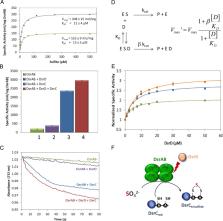
Dissimilatory sulfur metabolism was recently shown to be much more widespread among bacteria and archaea than previously believed. One of the key pathways involved is the dsr pathway that is responsible for sulfite reduction in sulfate-, sulfur-, thiosulfate-, and sulfite-reducing organisms, sulfur disproportionators and organosulfonate degraders, or for the production of sulfite in many photo- and chemotrophic sulfur-oxidizing prokaryotes. The key enzyme is DsrAB, the dissimilatory sulfite reductase, but a range of other Dsr proteins is involved, with different gene sets being present in organisms with a reductive or oxidative metabolism. The dsrD gene codes for a small protein of unknown function and has been widely used as a functional marker for reductive or disproportionating sulfur metabolism, although in some cases this has been disputed. Here, we present in vivo and in vitro studies showing that DsrD is a physiological partner of DsrAB and acts as an activator of its sulfite reduction activity. DsrD is expressed in respiratory but not in fermentative conditions and a Δ dsrD deletion strain could be obtained, indicating that its function is not essential. This strain grew less efficiently during sulfate and sulfite reduction. Organisms with the earliest forms of dsrAB lack the dsrD gene, revealing that its activating role arose later in evolution relative to dsrAB.
Related collections
Most cited references63
- Record: found
- Abstract: found
- Article: not found
Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC.
- Record: found
- Abstract: found
- Article: not found
Single cell activity reveals direct electron transfer in methanotrophic consortia.

- Record: found
- Abstract: found
- Article: found