- Record: found
- Abstract: found
- Article: found
Intact polar lipidome and membrane adaptations of microbial communities inhabiting serpentinite-hosted fluids
Read this article at
Abstract
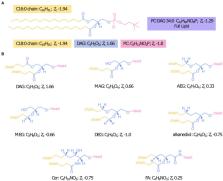
The generation of hydrogen and reduced carbon compounds during serpentinization provides sustained energy for microorganisms on Earth, and possibly on other extraterrestrial bodies (e.g., Mars, icy satellites). However, the geochemical conditions that arise from water-rock reaction also challenge the known limits of microbial physiology, such as hyperalkaline pH, limited electron acceptors and inorganic carbon. Because cell membranes act as a primary barrier between a cell and its environment, lipids are a vital component in microbial acclimation to challenging physicochemical conditions. To probe the diversity of cell membrane lipids produced in serpentinizing settings and identify membrane adaptations to this environment, we conducted the first comprehensive intact polar lipid (IPL) biomarker survey of microbial communities inhabiting the subsurface at a terrestrial site of serpentinization. We used an expansive, custom environmental lipid database that expands the application of targeted and untargeted lipodomics in the study of microbial and biogeochemical processes. IPLs extracted from serpentinite-hosted fluid communities were comprised of >90% isoprenoidal and non-isoprenoidal diether glycolipids likely produced by archaeal methanogens and sulfate-reducing bacteria. Phospholipids only constituted ~1% of the intact polar lipidome. In addition to abundant diether glycolipids, betaine and trimethylated-ornithine aminolipids and glycosphingolipids were also detected, indicating pervasive membrane modifications in response to phosphate limitation. The carbon oxidation state of IPL backbones was positively correlated with the reduction potential of fluids, which may signify an energy conservation strategy for lipid synthesis. Together, these data suggest microorganisms inhabiting serpentinites possess a unique combination of membrane adaptations that allow for their survival in polyextreme environments. The persistence of IPLs in fluids beyond the presence of their source organisms, as indicated by 16S rRNA genes and transcripts, is promising for the detection of extinct life in serpentinizing settings through lipid biomarker signatures. These data contribute new insights into the complexity of lipid structures generated in actively serpentinizing environments and provide valuable context to aid in the reconstruction of past microbial activity from fossil lipid records of terrestrial serpentinites and the search for biosignatures elsewhere in our solar system.
Related collections
Most cited references184
- Record: found
- Abstract: found
- Article: not found
DADA2: High resolution sample inference from Illumina amplicon data
- Record: found
- Abstract: not found
- Article: not found
Cutadapt removes adapter sequences from high-throughput sequencing reads

- Record: found
- Abstract: found
- Article: found