- Record: found
- Abstract: found
- Article: found
The evolution and genomic basis of beetle diversity
Read this article at
Significance
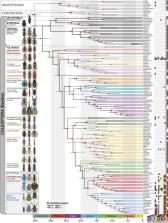
We inferred the phylogeny and evolution of beetles using genomic data of an unprecedented scale. Moreover, we documented the diversification of plant-feeding (herbivorous) beetles, which account for nearly half of all beetle species and a similar proportion of herbivorous insects, following convergent horizontal transfers of bacterial and fungal genes enabling the digestion of lignocellulose in plant cell walls. Our findings clarify beetle phylogenetic relationships and reveal new insights into the evolution of specialized herbivory and why there are so many species of beetles. Furthermore, they underscore the intimacy and complexity of the evolutionary relationships between insects, plants, and microorganisms and show how analyses of large-scale genomic data are revealing the evolution and genomic basis of insect biodiversity.
Abstract
The order Coleoptera (beetles) is arguably the most speciose group of animals, but the evolutionary history of beetles, including the impacts of plant feeding (herbivory) on beetle diversification, remain poorly understood. We inferred the phylogeny of beetles using 4,818 genes for 146 species, estimated timing and rates of beetle diversification using 89 genes for 521 species representing all major lineages and traced the evolution of beetle genes enabling symbiont-independent digestion of lignocellulose using 154 genomes or transcriptomes. Phylogenomic analyses of these uniquely comprehensive datasets resolved previously controversial beetle relationships, dated the origin of Coleoptera to the Carboniferous, and supported the codiversification of beetles and angiosperms. Moreover, plant cell wall-degrading enzymes (PCWDEs) obtained from bacteria and fungi via horizontal gene transfers may have been key to the Mesozoic diversification of herbivorous beetles—remarkably, both major independent origins of specialized herbivory in beetles coincide with the first appearances of an arsenal of PCWDEs encoded in their genomes. Furthermore, corresponding (Jurassic) diversification rate increases suggest that these novel genes triggered adaptive radiations that resulted in nearly half of all living beetle species. We propose that PCWDEs enabled efficient digestion of plant tissues, including lignocellulose in cell walls, facilitating the evolution of uniquely specialized plant-feeding habits, such as leaf mining and stem and wood boring. Beetle diversity thus appears to have resulted from multiple factors, including low extinction rates over a long evolutionary history, codiversification with angiosperms, and adaptive radiations of specialized herbivorous beetles following convergent horizontal transfers of microbial genes encoding PCWDEs.
Related collections
Most cited references62
- Record: found
- Abstract: not found
- Article: not found
Butterflies and Plants: A Study in Coevolution

- Record: found
- Abstract: found
- Article: found
SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data
- Record: found
- Abstract: not found
- Article: not found