- Record: found
- Abstract: found
- Article: found
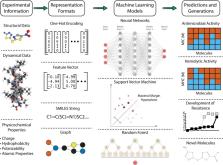
Accelerating antibiotic discovery through artificial intelligence
Read this article at
Abstract
By targeting invasive organisms, antibiotics insert themselves into the ancient struggle of the host-pathogen evolutionary arms race. As pathogens evolve tactics for evading antibiotics, therapies decline in efficacy and must be replaced, distinguishing antibiotics from most other forms of drug development. Together with a slow and expensive antibiotic development pipeline, the proliferation of drug-resistant pathogens drives urgent interest in computational methods that promise to expedite candidate discovery. Strides in artificial intelligence (AI) have encouraged its application to multiple dimensions of computer-aided drug design, with increasing application to antibiotic discovery. This review describes AI-facilitated advances in the discovery of both small molecule antibiotics and antimicrobial peptides. Beyond the essential prediction of antimicrobial activity, emphasis is also given to antimicrobial compound representation, determination of drug-likeness traits, antimicrobial resistance, and de novo molecular design. Given the urgency of the antimicrobial resistance crisis, we analyze uptake of open science best practices in AI-driven antibiotic discovery and argue for openness and reproducibility as a means of accelerating preclinical research. Finally, trends in the literature and areas for future inquiry are discussed, as artificially intelligent enhancements to drug discovery at large offer many opportunities for future applications in antibiotic development.
Abstract
Melo, Maasch and de la Fuente-Nunez review the current practices in use of artificial intelligence in the discovery of antibiotics and antimicrobials. They also provide details about the best-practices that should be engaged with during computational drug discovery, including open science and reproducibility.
Related collections
Most cited references169
- Record: found
- Abstract: found
- Article: not found
Long Short-Term Memory
- Record: found
- Abstract: found
- Article: found
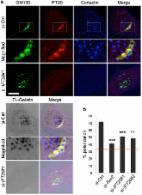
Ror2 signaling regulates Golgi structure and transport through IFT20 for tumor invasiveness

- Record: found
- Abstract: found
- Article: found