- Record: found
- Abstract: found
- Article: found
Complete mitochondrial genomes of two species of Stichopathes Brook, 1889 (Hexacorallia: Antipatharia: Antipathidae) from Rapa Nui (Easter Island)
research-article
Read this article at
There is no author summary for this article yet. Authors can add summaries to their articles on ScienceOpen to make them more accessible to a non-specialist audience.
Abstract
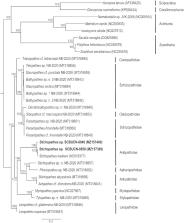
We report the complete mitochondrial genomes of two antipatharian species, Stichopathes sp. SCBUCN-8849 and Stichopathes sp. SCBUCN-8850, collected between 120 and 180 m depth off Rapa Nui (∼ −27.1°, −109.4°). The size of the two mitogenomes are 20,389 bp (29.0% A, 15.2% C, 19.9% G, and 35.9% T) and 20,463 bp (29.0% A, 15.3% C, 19.9% G, and 35.8% T), respectively. Both mitogenomes have the classic Hexacorallia gene content of 13 protein-coding, two rRNA, and two tRNA genes plus a COX1 intron with embedded HEG as found in the Antipathidae and other antipatharian families.
Related collections
Most cited references8

- Record: found
- Abstract: found
- Article: found
Trimmomatic: a flexible trimmer for Illumina sequence data
Anthony M. Bolger, Marc Lohse, Bjoern Usadel (2014)
- Record: found
- Abstract: found
- Article: not found
SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing.
Anton Bankevich, Sergey Nurk, Dmitry Antipov … (2012)
- Record: found
- Abstract: found
- Article: not found
MUSCLE: multiple sequence alignment with high accuracy and high throughput.
R. C. Edgar (2004)