- Record: found
- Abstract: found
- Article: found
Models in the Research Process of Psoriasis
Read this article at
Abstract
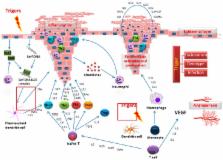
Psoriasis is an ancient, universal chronic skin disease with a significant geographical variability, with the lowest incidence rate at the equator, increasing towards the poles. Insights into the mechanisms responsible for psoriasis have generated an increasing number of druggable targets and molecular drugs. The development of relevant in vitro and in vivo models of psoriasis is now a priority and an important step towards its cure. In this review, we summarize the current cellular and animal systems suited to the study of psoriasis. We discuss the strengths and limitations of the various models and the lessons learned. We conclude that, so far, there is no one model that can meet all of the research needs. Therefore, the choice model system will depend on the questions being addressed.
Related collections
Most cited references84
- Record: found
- Abstract: found
- Article: not found
Mechanisms regulating skin immunity and inflammation.
- Record: found
- Abstract: found
- Article: not found
Integrative responses to IL-17 and TNF-α in human keratinocytes account for key inflammatory pathogenic circuits in psoriasis.
- Record: found
- Abstract: found
- Article: not found