- Record: found
- Abstract: found
- Article: not found
Exploring the active constituents of Oroxylum indicum in intervention of novel coronavirus (COVID-19) based on molecular docking method
Read this article at
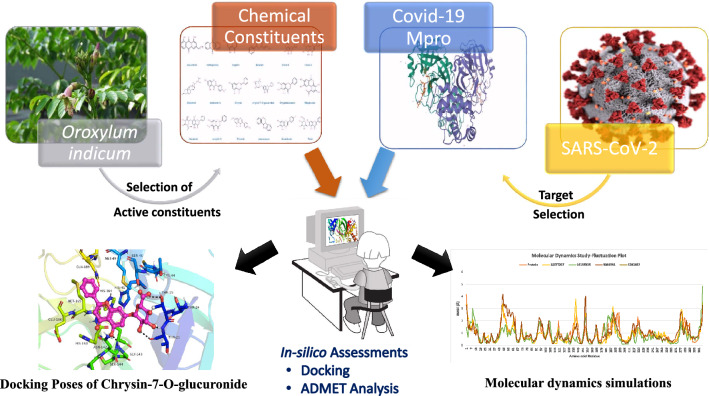
Graphical abstract
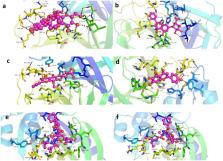
The severe acute respiratory syndrome COVID-19 declared a global pandemic by WHO has become the present wellbeing worry to the whole world. There is an emergent need to search for possible medications. We report in this study a molecular docking study of eighteen Oroxylum indicum molecules with the main protease (M pro) responsible for the replication of SARS-CoV-2 virus. The outcome of their molecular simulation and ADMET properties reveal four potential inhibitors of the enzyme (Baicalein-7- O-diglucoside, Chrysin-7- O-glucuronide, Oroxindin and Scutellarein) with preference of ligand Chrysin-7- O-glucuronide that has the second highest binding energy (− 8.6 kcal/mol) and fully obeys the Lipinski’s rule of five.
Related collections
Most cited references55
- Record: found
- Abstract: found
- Article: found
Open Babel: An open chemical toolbox
- Record: found
- Abstract: found
- Article: not found
Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China
- Record: found
- Abstract: found
- Article: not found
