- Record: found
- Abstract: found
- Article: found
Fragmented mitochondrial genomes of the rat lice, Polyplax asiatica and Polyplax spinulosa: intra-genus variation in fragmentation pattern and a possible link between the extent of fragmentation and the length of life cycle
Read this article at
Abstract
Background
Blood-sucking lice (suborder Anoplura) parasitize eutherian mammals with 67% of the 540 described species found on rodents. The five species of blood-sucking lice that infest humans and pigs have fragmented mitochondrial genomes and differ substantially in the extent of fragmentation. To understand whether, or not, any life-history factors are linked to such variation, we sequenced the mt genomes of Polyplax asiatica and Polyplax spinulosa, collected from the greater bandicoot rat, Bandicota indica, and the Asian house rat, Rattus tanezumi, respectively.
Results
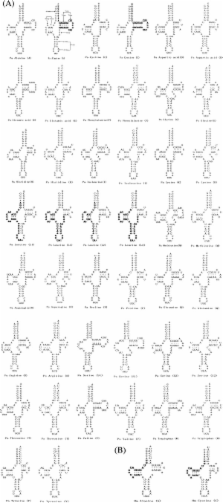
We identified all of the 37 mitochondrial genes common to animals in Polyplax asiatica and Polyplax spinulosa. The mitochondrial genes of these two rat lice are on 11 circular minichromosomes; each minichromosome is 2–4 kb long and has 2–7 genes. The two rat lice share the same pattern for the distribution of the protein-coding genes and ribosomal RNA genes over the minichromosomes, but differ in the pattern for the distribution of 8 of the 22 transfer RNA genes. The mitochondrial genomes of the Polyplax rat lice have 3.4 genes, on average, on each minichromosome and, thus, are less fragmented than those of the human lice (2.1 and 2.4 genes per minichromosome), but are more fragmented than those of the pig lice (4.1 genes per minichromosome).
Related collections
Most cited references15
- Record: found
- Abstract: found
- Article: not found
Identification of protein coding regions by database similarity search.
- Record: found
- Abstract: found
- Article: not found
The single mitochondrial chromosome typical of animals has evolved into 18 minichromosomes in the human body louse, Pediculus humanus.
- Record: found
- Abstract: found
- Article: not found