- Record: found
- Abstract: found
- Article: found
A new species of Munida Leach, 1820 (Crustacea: Decapoda: Anomura: Munididae) from seamounts of the Nazca-Desventuradas Marine Park
Read this article at
Abstract
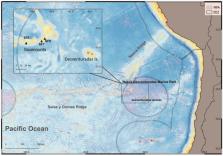
Munida diritas sp. nov. is described for the seamounts near Desventuradas Islands, in the intersection of the Salas & Gómez and Nazca Ridges, Chile. Specimens of the new species were collected in the summit (∼200 m depth) of one seamount and observed by ROV at two nearby ones. This species is characterized by the presence of distinct carinae on the thoracic sternites 6 and 7. Furthermore, it is not related with any species from the continental shelf nor the slope of America, while it is closely related to species of Munida from French Polynesia and the West-Pacific Ocean (i.e., M. ommata, M. psylla and M. rufiantennulata). In situ observations indicate that the species lives among the tentacles of ceriantarid anemones and preys on small crustaceans. The discovery of this new species adds to the knowledge of the highly endemic benthic fauna of seamounts of the newly created Nazca-Desventuradas Marine Park, emphasizing the relevance of this area for marine conservation.
Related collections
Most cited references47
- Record: found
- Abstract: found
- Article: not found
MUSCLE: multiple sequence alignment with high accuracy and high throughput.
- Record: found
- Abstract: found
- Article: found
Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data
- Record: found
- Abstract: found
- Article: not found