- Record: found
- Abstract: found
- Article: found
NMR solution structure of tricyclo-DNA containing duplexes: insight into enhanced thermal stability and nuclease resistance
Read this article at
Abstract
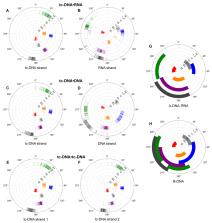
Tc-DNA is a conformationally constrained oligonucleotide analogue which shows significant increase in thermal stability when hybridized with RNA, DNA or tc-DNA. Remarkably, recent studies revealed that tc-DNA antisense oligonucleotides (AO) hold great promise for the treatment of Duchenne muscular dystrophy and spinal muscular atrophy. To date, no high-resolution structural data is available for fully modified tc-DNA duplexes and little is known about the origins of their enhanced thermal stability. Here, we report the structures of a fully modified tc-DNA oligonucleotide paired with either complementary RNA, DNA or tc-DNA. All three investigated duplexes maintain a right-handed helical structure with Watson-Crick base pairing and overall geometry intermediate between A- and B-type, but closer to A-type structures. All sugars of the tc-DNA and RNA residues adopt a North conformation whereas the DNA deoxyribose are found in a South-East-North conformation equilibrium. The conformation of the tc-DNA strand in the three determined structures is nearly identical and despite the different nature and local geometry of the complementary strand, the overall structures of the examined duplexes are very similar suggesting that the tc-DNA strand dominates the duplex structure.
Related collections
Most cited references41
- Record: found
- Abstract: found
- Article: not found
The R.E.D. tools: advances in RESP and ESP charge derivation and force field library building.
- Record: found
- Abstract: found
- Article: not found
LNA: a versatile tool for therapeutics and genomics.
- Record: found
- Abstract: found
- Article: not found